1 Introduction
papaja is short for ‘preparing APA journal articles’ and is the name of this R package designed to create fully reproducible journal articles that seamlessly fuse statistical analyses, simulations, and prose.
A manuscript written with papaja can be thought of as an extensively commented analysis script ready for publication in a scientific journal.
1.1 Motivation
APA style is one of the major style regimes in academia concerned with written scientific communication and is defined in the Publication Manual of the American Psychological Association [APA; American Psychological Association (2010)]. Research in psychology and many fields other, including other social sciences, medicine, and public health is reported in APA style. If you want to publish psychological research, you will have to produce properly formatted APA style manuscripts. While the merits of standardizing scientific reporting are undisputed, mastering the currently 272 pages of Publication Manual’s style definitions puts an undeniable burden on authors. To simplify the writing process researchers have developed strategies to automate the implementation of APA style.
The dominant approach to writing scientific reports in psychology is the use of common word processors such as Microsoft Word or Libre Office. Available APA style document templates set up page margins, line spacing, fonts, and related aspects of the manuscript. The format of citations and references can be automated by using references managers that integrate with the word processor, such as Zotero.
Far less common in psychology, is the use of markup systems, such as LaTeX or Markdown.
A key concept of markup systems is to separate style from content by means of document annotations.
These annotations declare portions of text for example as title, section headings, or list items but, crucially, they are agnostic to what this means visually (e.g., <emphasize>text</emphasize> instead of text).
There are several advantages to this approach but, crucially for the present purpose, markup systems compile plain text files to produce the final document.
Critically, the compilation process is required for fully reproducible scientific reports.
1.1.1 Reproducible scientific reports
Dynamic scientific reports fuse prose and simulation or analysis scripts (Gandrud, 2013; Xie, 2015). Each time a dynamic document is compiled, the scripts are executed anew and the results—statistics, tables, and figures—are recreated and inserted into the document. Thereby dynamic documents allow automated reporting of results, which is currently not possible with the prevalent use of Microsoft Word or Libre Office. Importantly, dynamic documents ensure that the results reported in the manuscript are free of rounding or transcription errors and correspond to the performed analyses. This is the reason why dynamic scientific reports are commonly referred to as reproducible scientific reports or reproducible reports. Thus, the term reproducibility here means that given a dataset and analysis script any analyst obtains the same statistical results (Asendorpf et al., 2013; Cacioppo, Kaplan, Krosnick, Olds, & Dean, 2015).
The importance of reproducibility was pinpointed by the U.S. National Science Foundation subcommittee on replicability in science:
Reproducibility is a minimum necessary condition for a finding to be believable and informative. (p. 4, Cacioppo, Kaplan, Krosnick, Olds, & Dean, 2015)
Good statistical practice is fundamentally based on transparent assumptions, repro-ducible results, and valid interpretations.
The ethical statistician […] strives to promote transparency in design, execution, and reporting or presenting of all analyses
Makes documentation suitable for replicate analyses, metadata studies, and other research by qualified investigators.
http://www.amstat.org/asa/files/pdfs/EthicalGuidelines.pdf
“an article about a computational result is advertising, not scholarship. The actual scholarship is the full software environment, code and data, that produced the result.” - Buckheit & Donoho, 1995
(Elsevier executable paper challenge: 10.1016/j.procs.2011.04.064)
Donoho, D. L., Maleki, A., Rahman, I. U., Shahram, M. & Stodden, V. Reproducible research in computational harmonic analysis. Comput. Sci. Eng. 11, 8–18 (2009). Donoho and fellow researchers have been at the forefront of reproducibility for many years; this a
org-mode: http://www.sciencedirect.com/science/article/pii/S0928425711000374?via%3Dihub http://yuyang0.github.io/static/doc/babel.pdf
Proprietary software also exists like Inference for R29 – for using R with Microsoft Office – but we do not have any experience with them. Open-source tools like Dexy30 and Sumatra31 are clearly very promising and have capabilities similar to Sweave and org-mode.
–>
R Markdown has been suggested as one possible framework for reproducible analyses (Allaire et al., 2017).
papaja is a R-package in the making including a R Markdown template that can be used with (or without) RStudio to produce documents, which conform to the American Psychological Association (APA) manuscript guidelines (6th Edition).
The package uses the document class apa6 and a .docx-reference file, so you can create PDF documents, or Word documents if you have to.
Moreover, papaja supplies R-functions that facilitate reporting results of your analyses in accordance with APA guidelines.
Markdown is a simple formatting syntax that can be used to author HTML, PDF, and MS Word documents (among others). In the following I will assume you have hopped onto the band wagon and know how to use R Markdown to conduct and comment your analyses. If this is not the case, I recommend you get to grips with R Markdown first. I use RStudio (which makes use of pandoc) to create my documents, but the general process works using any other text editor.
Although LaTeX is widely used to write entire manuscripts (not just equations), it is not commonly used in the field of psychology.
We suspect that the neglect is largely due to its complexity and shallow learning curve.
Both appear to outweigh the advantages of the system when it comes to handling citations and cross-references or typesetting large documents, complex tables, and equations.
Markdown paired with pandoc provides a simple interface to harness the power of LaTeX without requiring in-depth knowledge about LaTeX (see Document compilation).
1.2 Implementation
In this section we provide a quick overview of the software and the underlying design principles. Unless you are interested in these details it’s safe to skip this section.
1.2.1 Guiding principles
In our development of this package our design decisions are based on two guiding principles:
papajadocuments are convertible. A central motivation for the development of the package was the experience that many (if not most) scientific journals in psychology require that the final version of a submitted manuscript be provided as a Microsoft Word or RTF document. We, thus, strive to employ solutions that work for both LaTeX and Word output formats even if it means that some of the functions are less versatile than they could be.papajadoes not compute. The package is meant to facilitate writing reports and manuscripts in APA style; it is not meant to analyse data. Functions provided in this package do as little computing as possible to grant authors maximal analytic flexibility. Moreover, limiting the amount of computing done by the package facilitates package maintenance.
While we are guided by these principles, they are guidelines rather than commandments and we deviate from them if necessary.
For example, papaja functions do compute statistics that should be routinely reported according to APA guidelines, such as confidence intervals, if they are not provided in the analysis output objects.
We, however, try to enable customization and replacement of these defaults whenever we can.
1.2.2 Document compilation
It is important to note that papaja builds on several existing R packages, which in turn build on other software, to compile the final document.
This layered software design grants the package its capabilities but it comes at a cost:
When compilation of a papaja-document throws an error it may not be immediately obvious to an inexperienced user, which part of the process failed.
We, thus, provide an overview of the compilation process and the software involved.
The compilation process is outlined in Figure 1.1. As detailed in the R Markdown components section, an R Markdown document consists of three components: A YAML front matter that contains document meta data, prose written in markdown (and optionally LaTeX) syntax, and R code in so-called code chunks.
Additionally, the R code contained in an R Markdown document usually accesses data that are either stored in a file on the computers hard drive or a database on the local network or internet.
When the document is compiled either by calling rmarkdown::render() or by clicking the Knit-button in RStudio, the YAML front matter is parsed.
Errors during this processing step are usually due to erroneous YAML syntax and start with Error in yaml::yaml.load(enc2utf8(string), ...) :.
Next, the R code inside the R Markdown document is executed sequentially from top to bottom of the document and the resulting output is inserted into the document in markdown syntax.
Any figures that are generated during the execution of the R code are stored to the hard drive and embedded into the markdown document.
The R code is executed in a new R session with an empty workspace, no attached packages, and with the working directory set to the path of the R Markdown file.
If any portion of the R code throws an error the compilation is aborted.
In RStudio the debugger will point to the first line of the R code chunk that threw the error—not the exact line responsible for the error—and report the error message.
R error messages usually start with Error:, for example, Error: Object 'experiment_data' not found.
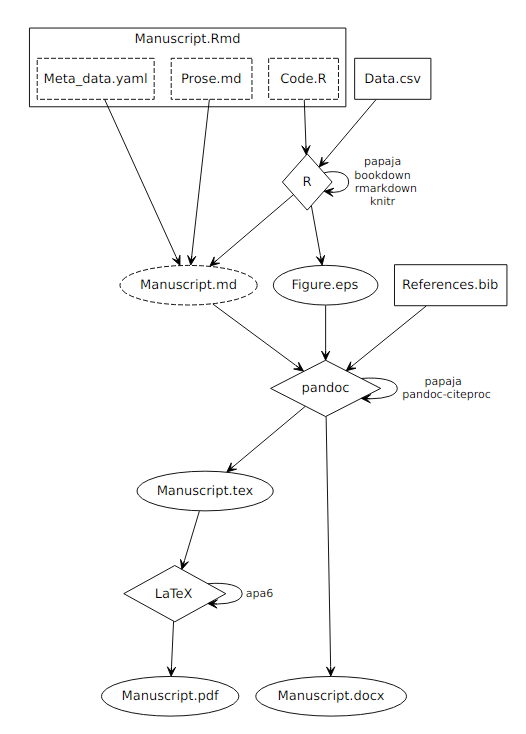
Figure 1.1: Process diagram illustrating the compilation of reproducible reports written with papaja. Boxes represent input files, diamonds represent compiling software, and ovals represent output files. Dashed components are either implicit (components of R Markdown files) or by default deleted after the compilation and, thus, invisible to the user.
After the R code has been executed the bookdown document format pdf_document2() or word_document2(), respectively, provides functions that enable the cross-referencing syntax \@ref() described in the Cross-referencing section.
Incorrect use of the cross-referencing syntax typically does not interfere with the document compilation but is apparent from missing or incorrect cross-references in the compiled document, e.g. instead of a figure number the document may contain the figure handle such as .
The result of this processing step is a pure markdown document containing only a YAML header, prose in markdown (and optionally LaTeX) syntax and rendered images in PDF, EPS, PNG, and TIFF formats at 300 dpi as set by apa6_pdf() or apa6_word(), respectively.
The markdown document, images and an optional BiB(La)TeX file that contains bibliographic information are the input to the next processing step.
rmarkdown::render() assembles a system call to pandoc that is supplemented by papaja to include a call to an additional filter and can be further customized by the user.
pandoc parses the contents of the markdown file and translates it into an abstract syntax tree (AST).
The pandoc-citeproc filter is than applied to the AST to replace the citation markup by the citation text and insert a reference section.
As detailed in the Citations section, pandoc-citeproc relies on the citation styles language (CSL), an XML-based specification of citation and bibliography formatting rules.
Unfortunately, CSL does not support in-text citations.
To overcome this limitation, pandoc-citeproc provides a separate syntax for in-text citations, which is based on the same CSL style.
As a result the CSL APA6 style uses ampersands for all citations, including in-text citations, defying the APA citation style.
That is why papaja provides an additional R-based filter that is applied to the AST to replace ampersands in in-text citations with the word and.
After all filters have been applied to the AST pandoc converts the document into the target output format, that is, PDF or DOCX.
To create DOCX-documents papaja provides a reference DOCX-file containing page setup and style definitions that pandoc uses to create the manuscript file.
That is, pandoc translates the AST directly to the office open XML format.
To create PDF-documents pandoc in turn relies on LaTeX, that is, pandoc translates the AST to a TeX-file based on a template provided by papaja.
This template invokes the apa6 LaTeX document class (citation_needed?) and loads several additional LaTeX packages.
pandoc then generates a system call to LaTeX, which renders the PDF document.
During this processing step, that is, following the pandoc system calls, errors are due to failures of LaTeX to compile the TeX-file and usually start with !, for example, ! Undefined control sequence. or ! Missing $ inserted.
We provide some advice on Troubleshooting later in this manual.
1.3 Alternatives
Needless to say, not all journals require manuscripts to be prepared according to APA guidelines.
Other journal templates for the rmarkdown package are available in the rticles package.
It includes a set of templates for authoring of R related journal and conference submissions.
- papeR
- reportR & apaStyle
- Sweave
- schoRsch (Pfister & Janczyk, 2016)
- sigr
1.4 Getting started
1.4.1 Software requirements
papaja depends on additional software, namely,
If you work with RStudio (1.1.453 or later) pandoc should already be installed, otherwise refer to the installation instructions for your operating system.
1.4.1.1 Setting up a TeX distribution
If you want to create PDF- in addition to DOCX-documents you additionally need
- TeX 2013 or later.
If you have no use for TeX beyond rendering R Markdown documents, I recommend you use TinyTex. Otherwise consider MikTeX for Windows, MacTeX for Mac, or TeX Live for Linux. TinyTex can be installed from within R as follows.
if(!"tinytex" %in% rownames(installed.packages())) install.packages("tinytex")
tinytex::install_tinytex()papaja requires additional \(\LaTeX\) packages that are not part of the basic TeX installations.
When using TinyTex, the required packages will be installed automatically when rendering a the first PDF-document.
Users of other TeX distribution need to take one of the following additional steps.
MikTeX users may enable the automatic installation of missing packages.
If you are comfortable using the command line, it may be convenient to run the following command.
initexmf --set-config-value [MPM]AutoInstall=1This option is not available in MacTeX or TeX Live. Alternatively, you can install the additional packages manually. Again the installation can be done conveniently from the command line. MikTeX users can run the following command.
mpm --install=apa6,booktabs,caption,csquotes,endfloat,environ,etoolbox,fancyhdr,fancyvrb,framed,lineno,microtype,mptopdf,ms,parskip,pgf,sttools,threeparttable,threeparttablex,trimspaces,txfonts,upquote,url,was,xcolorIt may be necessary to launch MikTeX console as administrator (right-click and Run as administrator) before executing the command.
TeXLive and MacTeX users can run the following command.
tlmgr install apa6 booktabs caption csquotes endfloat environ etoolbox fancyhdr fancyvrb framed lineno microtype mptopdf ms parskip pgf sttools threeparttable threeparttablex trimspaces txfonts upquote url was xcolorIt may be necessary to first install tlmgr.
Alternatively, Ubuntu users can install the following Ubuntu packages:
sudo apt-get install texlive-publishers texlive-fonts-extra texlive-latex-extra texlive-humanities lmodernFinally, you may install a complete—not the basic—TeX distribution that comes with all available LaTeX packages but is several gigabytes large.
1.4.2 Installing papaja
papaja is not yet available on CRAN but you can install it from GitHub:
# Install devtools package if necessary
if(!"devtools" %in% rownames(installed.packages())) install.packages("devtools")
# Install the stable development verions from GitHub
devtools::install_github("crsh/papaja")
# Install the latest development snapshot from GitHub
devtools::install_github("crsh/papaja@devel")papaja has not yet been submitted to CRAN because it is under active development.
Currently, there are still a couple of loose ends we would like to tie up before we release the package to a larger audience.
If you would like to contribute to speed up the process, have a look at the chapters Limitations and Future directions.
1.4.3 Creating a document
Once you have installed papaja and all other required software, the APA manuscript template is available through the RStudio menus when creating a new R Markdown file, Figure 1.3.
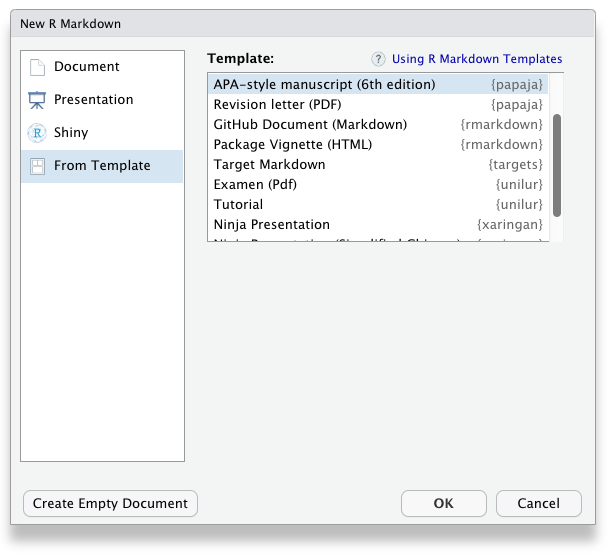
Figure 1.3: papaja’s APA6 template is available through the RStudio menues.
When you click RStudio’s Knit button (Figure 1.4) papaja, rmarkdown, and knitr work together to create an APA conform manuscript that includes both your manuscript text and the results of any embedded R code.
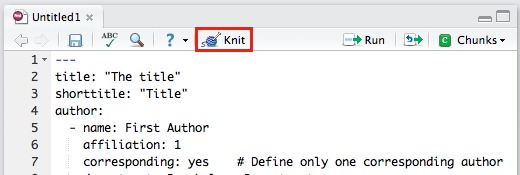
Figure 1.4: The Knit button in the RStudio.
If you don’t use RStudio, you can achieve the same result via the R command line.
Use rmarkdown::draft() to create a new R Markdown file and rmarkdown::render() to render the document.
# Create new R Markdown file
rmarkdown::draft(
"mymanuscript.Rmd"
, "apa6"
, package = "papaja"
, create_dir = FALSE
, edit = FALSE
)
# Render manuscript
rmarkdown::render("mymanuscript.Rmd")papaja is a R-package in the making including a R Markdown template that can be used with (or without) RStudio to produce documents, which conform to the American Psychological Association (APA) manuscript guidelines (6th Edition). The package uses the LaTeX document class apa6 and a .docx-reference file, so you can create PDF documents, or Word documents if you have to. Moreover, papaja supplies R-functions that facilitate reporting results of your analyses in accordance with APA guidelines.